Welcome to the universe of non-coding RNAs! This is a gigantic realm – our cells transcribe over 75% of the genome into RNAs that are never translated! [1, 2] – where the unknown is not just around the proverbial corner, but it often stares you straight in the eyes… Over the next few weeks, I’ll be your humble guide in this journey through the puzzling, mysterious realm of ncRNAs.
In this first stop, we will discuss the two types of ncRNAs at the heart of translation, and a third one, definitely more mysterious.
Let’s get going fellow travellers!
1. ribosomal RNA (rRNA)
The most abundant RNA in the cell – they make up to 85% of all transcripts [3].
rRNAs are essential components of the ribosome, the macromolecular machinery that synthesises new proteins. In fact, rRNAs
- are the structural backbone of the ribosome,
- stabilise the tRNAs loaded with their amino-acid residues,
- form the peptide bond linking amino acid residues in the growing polypeptide chain.
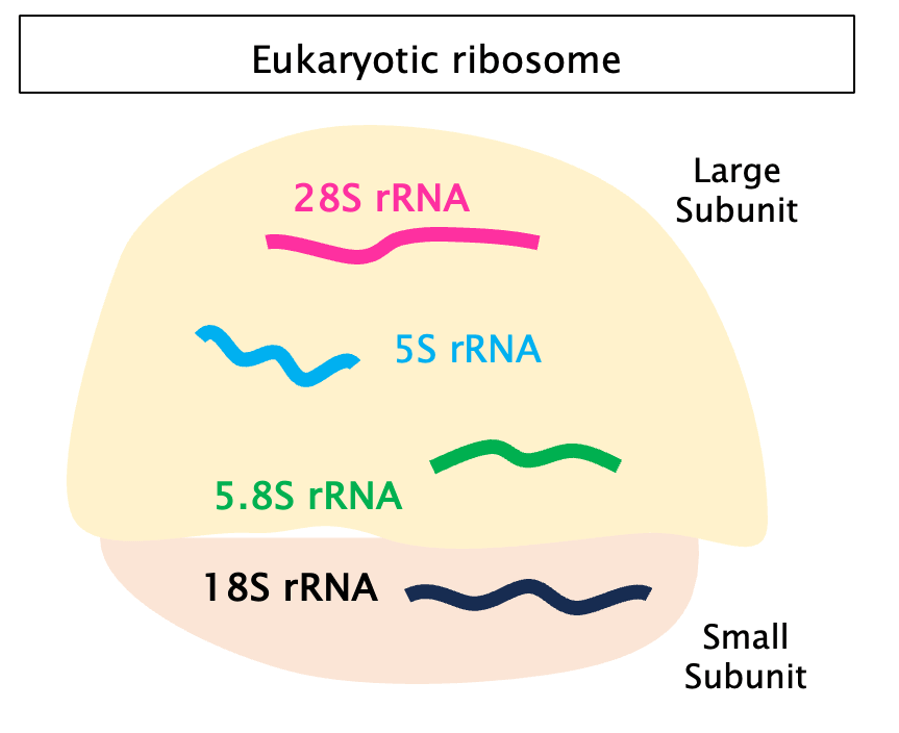
Eukaryotes have 4 types of rRNA: 3 in the large (60S) and one in the small (40S) ribosomal subunit. Instead, prokaryotes have 3: 2 in the large (50S) and one in the small (30S).
In humans, rRNAs range from ~120 (5S rRNA) to ~1900 bp (18S rRNA).
2. transfer RNA (tRNA)
The second most abundant RNA in the cell – they make up the 10-12% of all transcripts [3]. They act as adapters between mRNA and proteins: they match codons (mRNA triplets) with the corresponding amino acids in the ribosome.
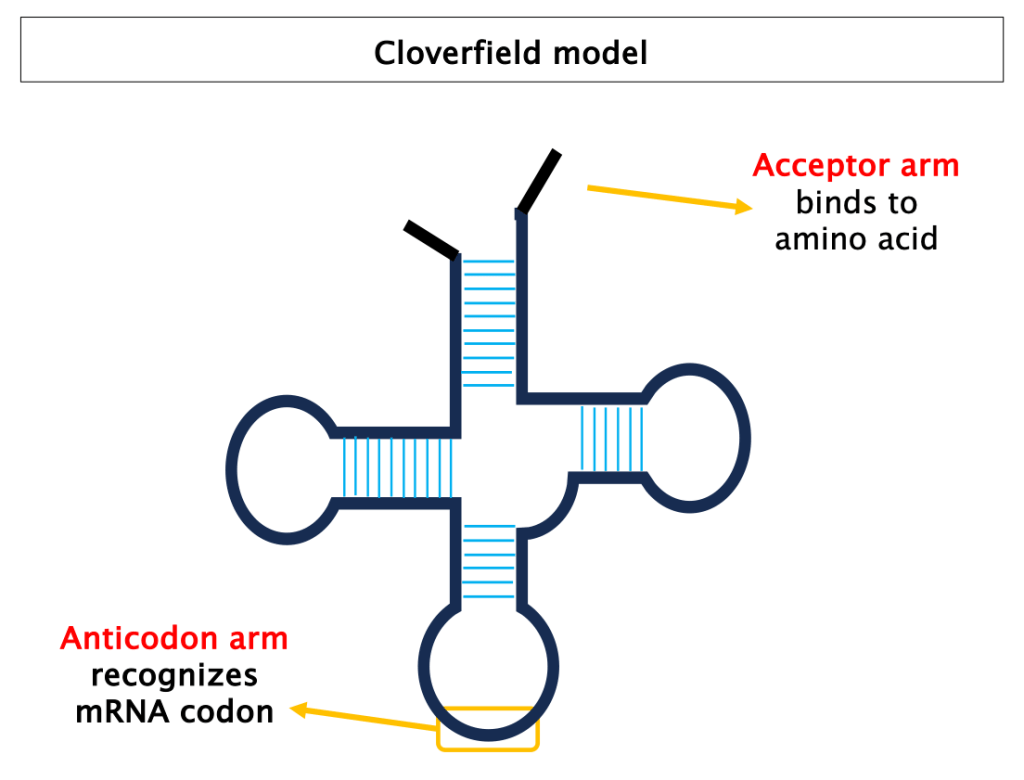
tRNAs are molecules between 76 and 90 nucleotides long. They share an universal secondary structure, known as cloverleaf, which was first proposed in 1965 (read here the story of how the first tRNA was sequenced). In 3D, tRNAs fold into an L-shaped structure: the anticodon arm anneals with mRNAs codons, the acceptor arm associates to a specific amino acid.
The human genome harbours over 600 tRNA genes [4], which are transcribed in precursor-tRNAs (pre-tRNAs) and undergo maturation in the nucleus. Sometimes, tRNAs and pre-tRNAs undergo alternative cleavages, which result in the production of tsRNAs.
3. transfer-derived small RNA (tsRNA)
They are derived from the cleavage of tRNAs and pre-tRNAs by angiogenin, Dicer or possibly other RNases. The biological functions of tsRNAs are still under investigation.
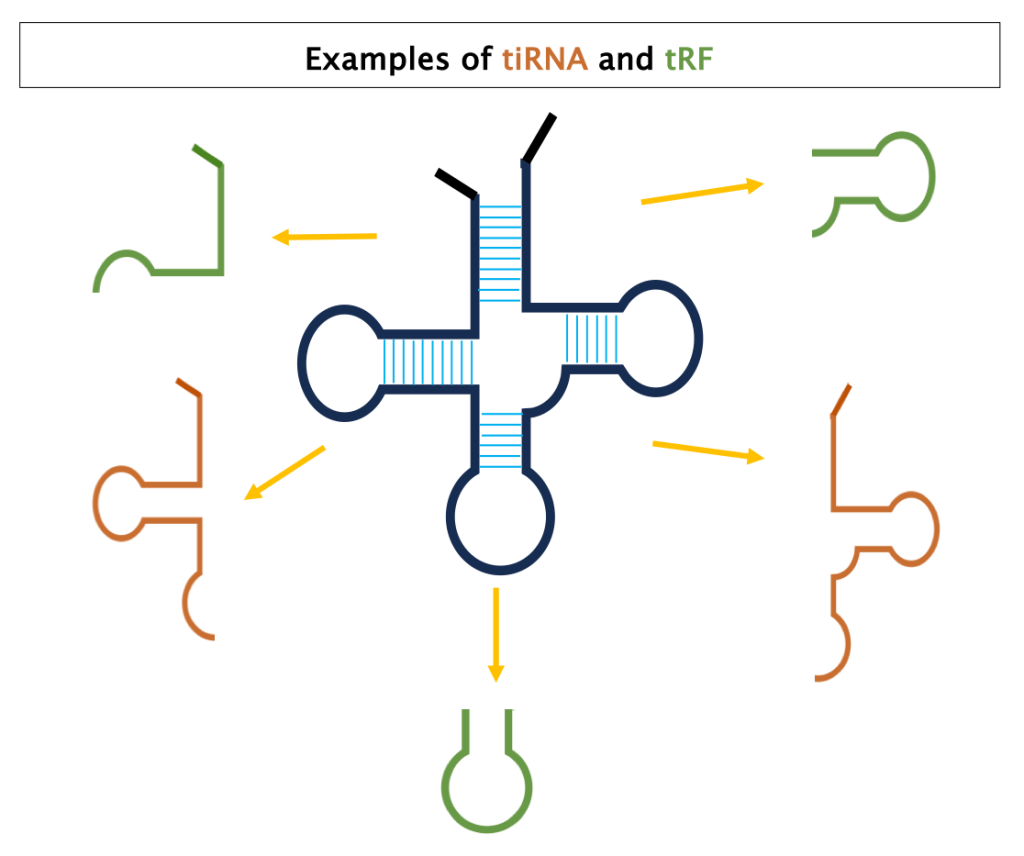
The cleavage can occur at different positions of the tRNA and pre-tRNAs, releasing two major classes of tsRNA: tRNA halves (tiRNAs) and tRNA-derived fragments (TRFs). Both tiRNAs and tRF have multiple subclasses ranging from 14 to 40 nt in length [5].
tdRNAs can suppress gene expression via RNA interference (RNAi) like miRNAs do. In addition tdRNA can regulate protein synthesis via association to rRNA and ribosomal protein subunits.
WHERE ARE WE GOING NEXT?
In the next episode, we pay a visit to three new types of ncRNA:
- small nucleolar RNA (snoRNA)
- sno-derived RNA (sdRNA)
- small Cajal body RNA (scaRNA)
REFERENCES
- Poliseno L et al. Coding, or non-coding, that is the question. Cell Res 34, 609–629 (2024). https://doi.org/10.1038/s41422-024-00975-8.
- Piovesan et al. Human protein-coding genes and gene feature statistics in 2019 (2019) BMC Res Notes.
- Bionumbers, Harvard
- Pan. Modifications and functional genomics of human transfer RNA (2018) Cell Res
- Shen et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases (2018) J Mol Med 96, 1167–1176.

Leave a reply to How many types of ncRNA do you know? snoRNA, sdRNA, scaRNA (Ep. 2) – WritinGenomics Cancel reply