This week, our journey in the ncRNAs universe brings us back to miRNAs before moving to splicing and the RNAs that make it possible.
Let’s get going fellow travellers!
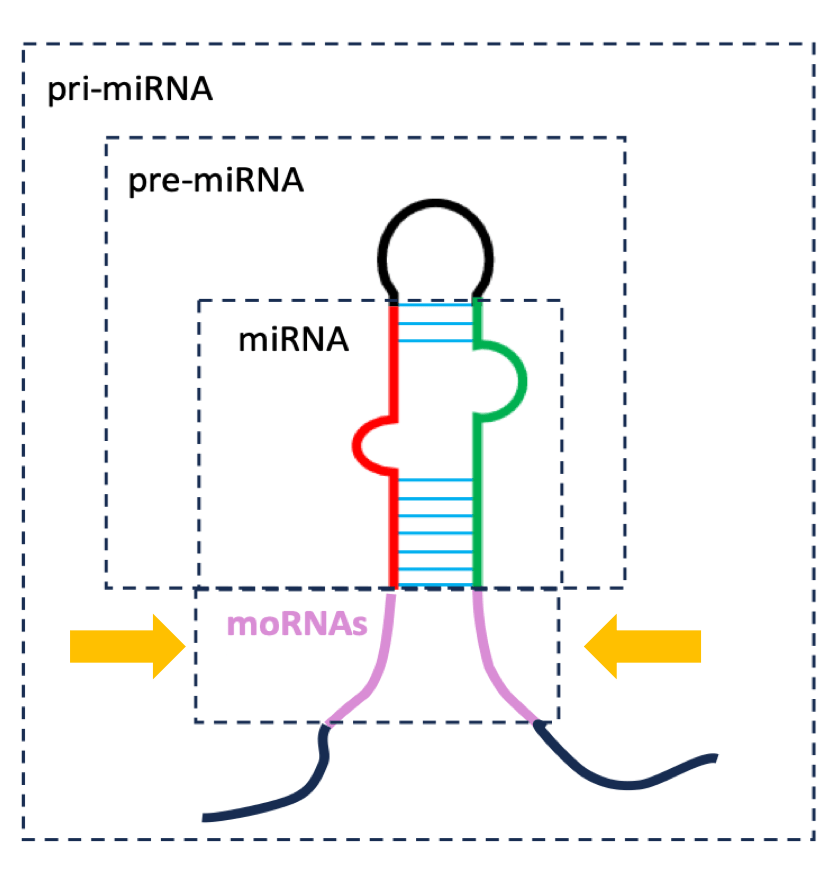
10. microRNA-offset RNA (moRNA)
These ~ 20 nt long transcripts are coded in the regions of the pri-miRNA directly flanking the pre-miRNA.
They were initially considered by-products of miRNA processing, but several lines of evidence suggest moRNA may be biologically active, and not merely “junk”. For instance,
- The biogenesis of moRNAs likely depends on a dedicated pathway, distinct from the one used by miRNAs [1];
- moRNA levels do not correlate with their cognate miRNA – some moRNAs are more abundant than the adjacent miRNAs [1, 2];
- moRNAs are evolutionary conserved, when their cognate miRNA is [1],
- moRNA levels appear to be regulated by stage of development and cell type [2].
At least one moRNA is biologically active (in mice, at least). I am referring to moRNA-21, which alters gene expression and promotes the proliferation of vascular smooth muscle cells [3].
11. circular RNA (circRNA)
These are single strand RNAs looped onto themselves, ranging from 100 to several thousand nucleotides. These molecules regulate gene expression by sequestering miRNAs. In a nutshell, they promote gene expression through a double negative: they inhibit ncRNAs that silence genes.
CircRNAs are generated in the nucleus from pre-mRNAs through a type of alternative splicing known as back-splicing. In this process, the 5’ of an exon is covalently bound to the 3’ of the same exon or of an exon upstream. The linear and circular form of a pre-mRNA are in competition. Back-splicing can occur through multiple mechanisms and can yield different types of circRNA, containing only exons, or exons and introns. CircRNAs made out of an intron do exist as well, but they are generated by mechanisms distinct from back-splicing.
Exon-containing circRNAs are usually exported in the cytoplasm. There, they bind to miRNAs – circRNAs harbour multiple miRNA-responsive elements. This mechanism reduces the miRNAs available and, as a result, reduces their inhibition on target mRNA, whose expression therefore increases. In virtue of their ability to bind and remove miRNA – you may say sucking them up – circRNAs are called “miRNA sponges”.
Despite their classification as non-coding RNAs, some exonic circRNAs are indeed translated.

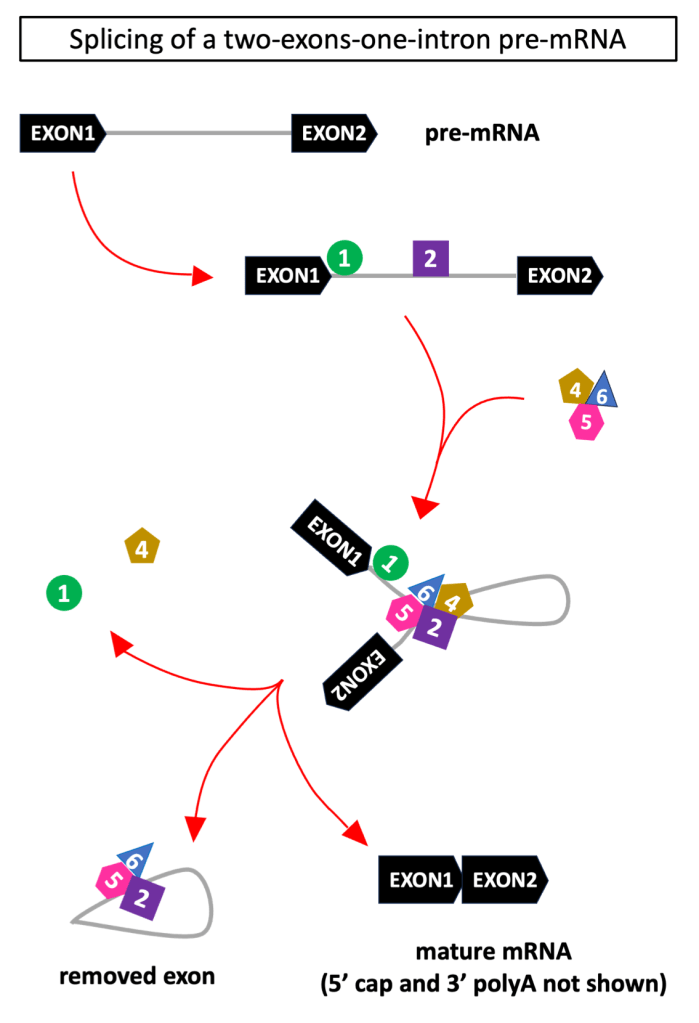
12. small nuclear RNA (snRNA)
They are transcripts shorter than 200 nucleotides. Each snRNA assembles with a protein into a ribonucleoprotein (RNP). Their main function is to orchestrate splicing, that is joining of exons wile removing introns in a pre-mRNA (don’t fall into the most common misconception surrounding exons!)
RNPs assemble in two complexes: the major spliceosome (formed by U1, U2, U4, U5, U6) and minor spliceosome (U11, U12, U4atac, U5, U6atac). The major complex intervenes in most instances, while the minor complex handles the removal of a rare (<1%) type of intron.
In these RNPs, snRNAs play 3 major roles:
- structural support: they are the scaffold of RNPs;
- guide: they direct the RNPs onto the sequences of the pre-mRNA to be cut and pasted;
- catalytic unit: they catalyse the biochemical reactions transforming the pre-mRNA into a mRNA.
WHERE ARE WE GOING NEXT?
In the next episode, we pay a visit to three new types of ncRNA:
- Y RNA
- vault RNA (vRNA)
- extracellular (exRNA).
REFERENCES
- Langenberger et al. Evidence for human microRNA-offset RNAs in small RNA sequencing data (2009) Bioinformatics.
- Asikainen et al. Selective MicroRNA-Offset RNA Expression in Human Embryonic Stem Cells (2015) PLOS One.
- Zhao et al. MicroRNA-Offset RNA Alters Gene Expression and Cell Proliferation (2016) PLOS One.
- Jouravleva, Zamore. A guide to the biogenesis and functions of endogenous small non-coding RNAs in animals (2025) Nature Reviews Molecular Cell Biology.

Leave a comment